
I graduated from the University of Cambridge in 1970 and obtained a PhD at the University of Manchester in 1974. With Struther Arnott at Purdue University, I used fibre diffraction to determine the structures of DNA and RNA polynucleotides. In 1977, I joined Michael Rossmann’s group at Purdue for the successful determination of the structure of Southern Bean Mosaic Virus capsid. I returned to the UK in 1979 to the Biophysics Group at Imperial College London, led by Professor David Blow. While at Imperial, I determined the structures of glyceraldehyde 3-phosphate dehydrogenase in various states of cofactor ligation in a collaboration with Alan Wonacott, and the structure of chloramphenicol acetyltransferase (CAT) in collaboration with Bill Shaw from the University of Leicester.
I joined the LMB in 1988, initially continuing studies on CAT but also collaborating with Robin Carrell (Haematology) on the structure of ovalbumin as a model for an uncleaved serine protease inhibitor (SERPIN). At this point John Walker, who was initially PNAC and then the Dunn Nutrition Unit, obtained the first well-diffracting crystals of the soluble (F1) component of ATP synthase. In a collaboration that continued for almost 30 years, the crystal structures of bovine F1-ATPase trapped in various stages of the catalytic cycle gave considerable insight into the remarkable rotary catalytic mechanism of ATP hydrolysis and synthesis. Complexes with the natural inhibitor protein, the peripheral stalk proteins and the membrane-associated c-ring provided a more complete visualisation of this complex assembly. The crystal structures of several bacterial F1-ATPases were also determined with a view to aiding antibiotic design, culminating in the 4 Å resolution structure of an intact bacterial ATP synthase.
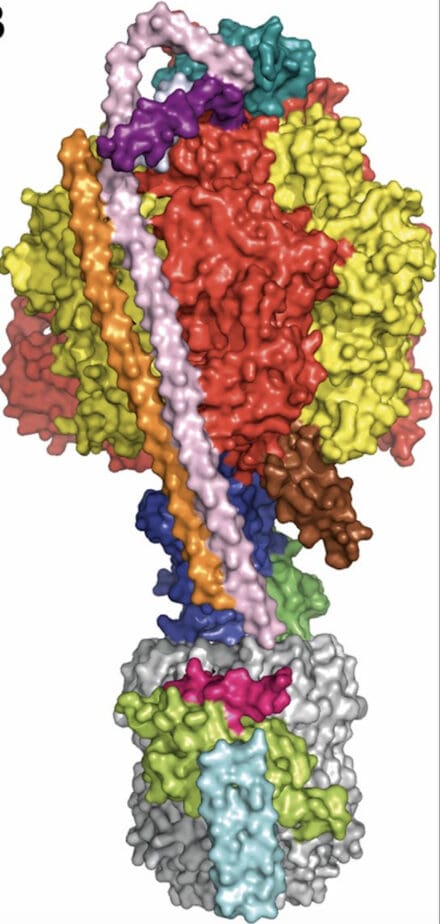
Using cryo-EM techniques, Tony Crowther generated a 6 Å resolution model of the Hepatitis B capsid. This was used as a phasing model to solve the atomic structure of the capsid. A breakthrough thermostabilisation technique developed by Chris Tate led to the structure determination of several G protein-coupled receptors, in both the activated (agonist bound) and inactive (antagonist bound) states, explaining the mechanism of activation. This was enhanced by the structure of the A2A receptor in complex with a minimal G protein construct.
The data processing software iMOSFLM was also extensively developed while I was working at the LMB.