Sjors Scheres
Structural biology of neurodegeneration

Almost all cases of neurodegeneration are characterised by the abundant presence of filamentous protein aggregates, or amyloids, in the brain. But how amyloid filaments form in the brain and how their presence relates to disease remains a mystery.
We develop new image processing methods for atomic structure determination of biological macromolecules by cryo-EM and cryo-ET. We implement these methods in our open-source computer programme RELION, which is used by cryo-EM labs all over the world.
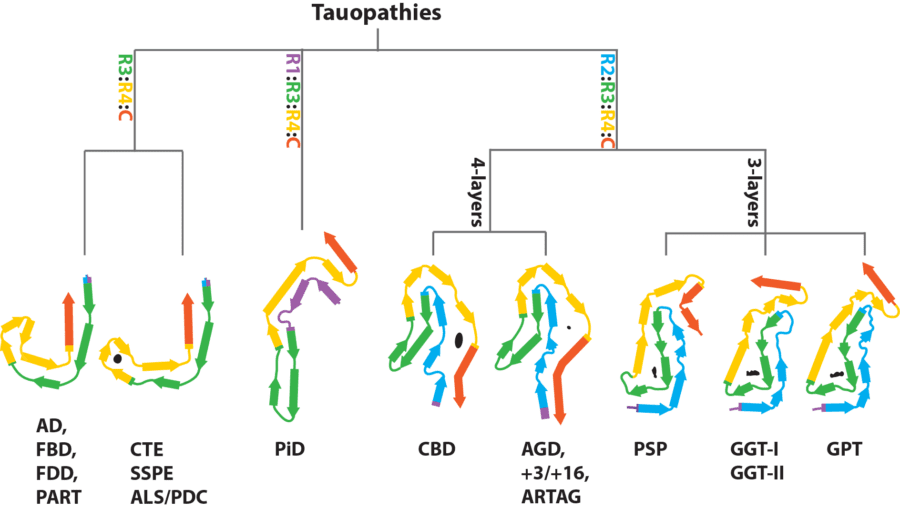
Our methods unlocked the possibility to study the atomic structures of amyloid filaments, and we now use these methods on filaments from human brains with neurodegenerative disease. This work has shown that a given protein may adopt many different amyloid structures. For example, tau forms filaments in more than 20 neurodegenerative diseases and our work has revealed that different tau folds characterise distinct diseases. Our ongoing efforts aim to understand how and why this happens. This line of research is carried out in close collaboration with Michel Goedert’s group.