The method is based on de novo design of protein biomaterials and enables polarisation of cells with any protein of interest
Asymmetric cell division is the process by which one cell divides into two daughter cells that have different fates. It is the hallmark of stem cells, in which one daughter cell specialises to perform a specific function in the organism, while the other becomes another stem cell that keeps the ability to divide. However, dissection of the numerous polarity pathways that act upon asymmetric cell division in multicellular organisms has proven challenging because they often act simultaneously and thus cannot be untangled. Now Emmanuel Derivery’s group, in the LMB’s Cell Biology Division, has developed a method to directly untangle the contribution of any signalling pathway involved in this process by artificially polarizing normally unpolarized cells. Subsequently, using this method shed new light on the mechanisms of cytoskeleton symmetry breaking, a crucial aspect of asymmetric cell division.
In this study, co-led by Joseph Watson and Lara Krüger, the team developed an assay which allows the contribution of each pathway involved in the process to be dissected. The assay is based on de novo designed proteins that assemble into 2D lattices that were previously developed by the group in collaboration with David Baker’s lab at the University of Washington.
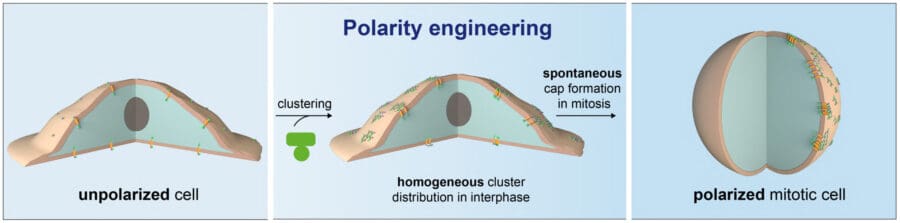
Joseph, former member of Emmanuel’s group and now postdoc at the University of Washington, demonstrated that the designed protein material spontaneously forms “caps” at the surface of dividing cells and thus polarizes the normally unpolarized culture cells. This is achieved by clustering a transmembrane domain from the outside using the material, to which the intracellular protein of interest is targeted. These clusters coalesce and form asymmetric “caps” when cells round up in division therefore polarizing the cell with the protein of interest (see image above).
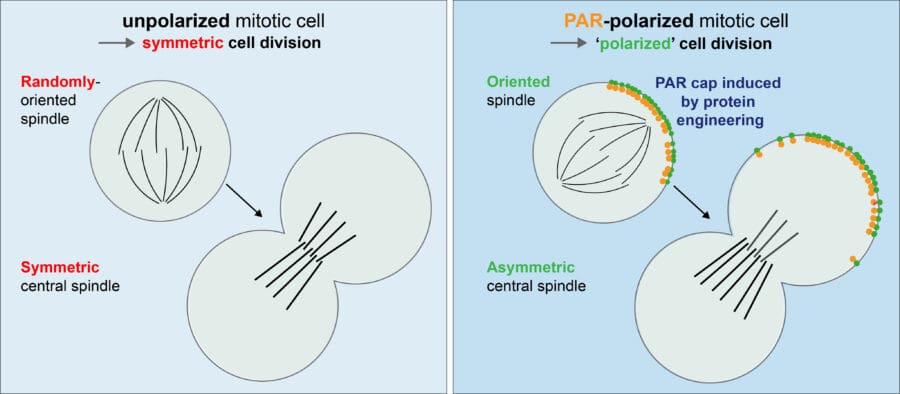
He then demonstrated that artificial caps of the Par-complex, a key regulator of asymmetric cell division, induces two hallmarks of asymmetric cell division, namely spindle orientation along the established polarity axis and central spindle symmetry breaking. Hence, a Par complex polar cap results in a “polarized” division in a cell that would otherwise divide symmetrically (see image below).
Lara then used the assay to unravel the molecular mechanisms by which an asymmetric Par complex cap can induce these hallmarks of asymmetric cell division. Specifically, she showed that these hallmarks occur via different pathways that can be untangled and identified the molecular cascade of proteins involved.
James Manton, in the LMB’s Cell Biology Division, used the extraordinary speed of the subcellular light sheet microscope to unravel how the polymer forms caps at the surface of dividing cells. The reason turned out to be geometrical, not biological: the polymer spontaneously makes caps on rounded-up cells, which happens in mitosis. Marta Shahbazi from the LMB’s Cell Biology Division confirmed that the method was applicable in other cell types by demonstrating it could apply to reconstitute polarity in mouse embryonic stem cells.
It has never been possible to reconstitute as many of the pathways governing asymmetric cell division as this new method allows. Scientists can now polarize cells with any protein of interest and follow cells through division to investigate the downstream and later occurring effects of these polarity cues. The method also directly permits high-speed, high-resolution imaging of these phenomena. Altogether, this permits the discovery of new biology. For example, in this work it was discovered that an asymmetric central spindle, which is known to be crucial in asymmetrically dividing fly cells, is also present in asymmetrically dividing mammalian cells.
More broadly, defects in asymmetric cell division have been implicated in diseases such as cancer. This method may eventually help to determine “what goes wrong” when stem cell related cancers or other diseases develop, and thus inspire the design of targeted treatments.
This work was funded by UKRI MRC, the Human Frontier Science Program, EMBO and the Wellcome Trust.
Further references
Synthetic Par polarity induces cytoskeleton asymmetry in unpolarized mammalian cells. Watson, JL., Krüger, LK., Ben-Sasson, AJ., Bittleston, A., Shahbazi, M., Planelles-Herrero, VJ., Chambers, JE., Manton, JD., Baker, D., Derivery, E. Cell
Emmanuel’s group page


