Suyang Zhang
Mechanism of transcription-coupled alternative splicing

Splicing of precursor messenger RNA (pre-mRNA) is a fundamental step in eukaryotic gene expression. Within the cell, splicing occurs simultaneously with pre-mRNA synthesis by RNA polymerase II (Pol II) and is mechanistically coupled to transcription. The tightly coordinated crosstalk between the transcription and splicing machineries enhances the fidelity of gene expression and is crucial for cell survival and functionality. More than 95% of human genes are alternatively spliced, massively expanding the coding potential of our genome. Dysregulation of alternative splicing contributes to cancer pathogenesis and is being investigated as a potential biomarker for disease diagnosis and prognosis.
Our research aims to understand the mechanism of gene regulation in a physiologically relevant context, focussing on how transcription, splicing and chromatin structure collectively shape gene expression. We employ a multidisciplinary approach, combining structural biology, biochemical reconstitution, functional assays and in vivo RNA sequencing.
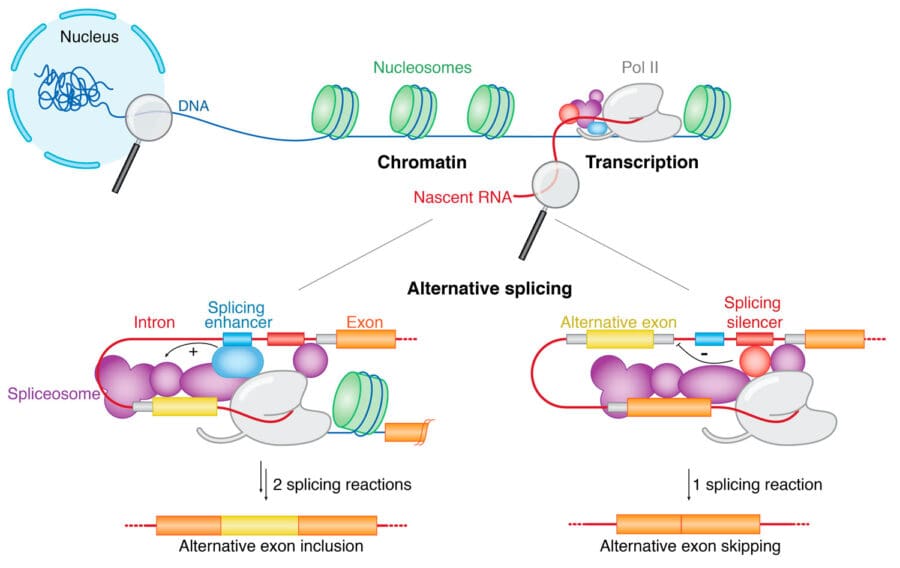
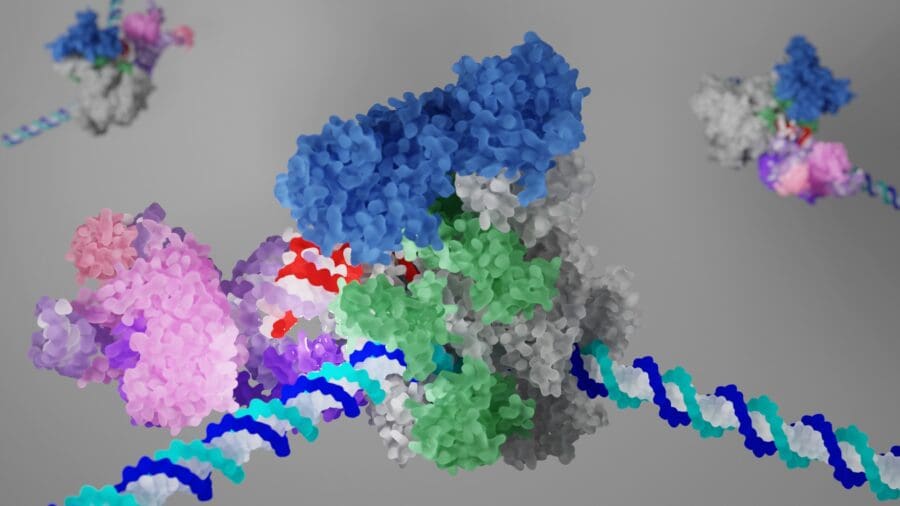
Transcription and splicing are coordinated by molecular interactions and the rate of Pol II elongation. Our cryo-EM structure of the elongating Pol II in complex with U1 snRNP, the first spliceosome component recruited to the pre-mRNA, revealed for the first time direct protein-protein interactions between the transcription and splicing machineries. This provided crucial insights into how functional pairing of distant intron ends and spliceosome assembly can occur on the Pol II surface. Furthermore, we identified a regulatory mechanism that modulates Pol II elongation rate, potentially facilitating the coordination of co-transcriptional processes. Our long-term goal is to provide fundamental insights into how transcription, splicing and chromatin structure are coordinated to regulate gene expression.