
Our group focusses on the development of mathematical, statistical and computational techniques to derive reliable atomic models from noisy and incomplete experimental data, with a focus on automation. We employ Bayesian statistical frameworks to integrate data from multiple sources, including stereochemical and structural prior knowledge, as well as experimental data obtained from crystallography and cryo‑EM.
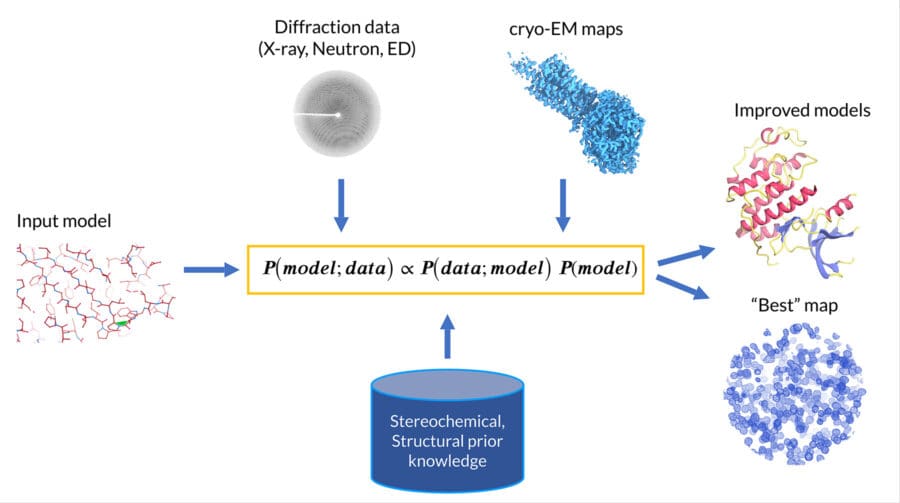
Our group has developed some of the widely used software tools among computational structural biologists. These include REFMAC5 for atomic model refinement and updated map generation; AceDRG for the organisation and application of stereochemical restraints; ProSMART for the alignment and superposition of macromolecules, and restraint generation; EMDA for map manipulation and maximum likelihood map averaging; LIBG for restraint generation for nucleic acid base pairing and stacking; Servalcat for atomic structure model refinement and map calculations; and MetalCoord to enhance the restraining, refinement and validation of structures containing metal ligands.
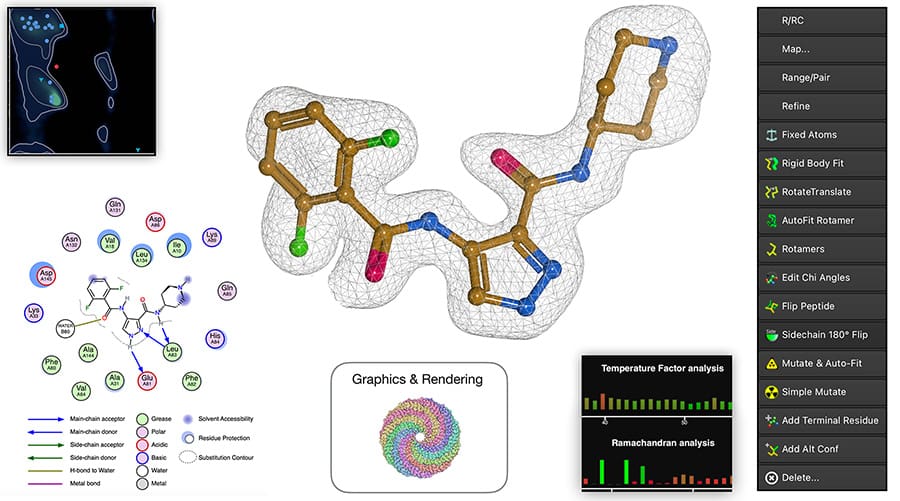
Paul Emsley leads an international team working on Coot, a widely used tool for building, refining, visualising and validating atomic models. Recent enhancements to Coot include: improved random-start rigid-body refinement with a larger radius of convergence; tools for low-resolution refinement; the implementation of RNA/DNA restraints; enhanced building and refinement of N-linked glycosylations; and integration with the AceDRG interface for ligand linkages and refinement of metal coordination. In addition, his group is actively developing Moorhen, a web-based model-building and refinement tool. This software is designed to be accessible from virtually any location and on any device.