
Synthetic biology enables us to control certain genetic programmes and introduce novel phenotypes in model bacteria such as E. coli, though as a field we cannot comprehensively engineer the bacteria that significantly affect human health. Our group develops genome engineering tools for the de novo synthesis of non-model gut bacteria, with the ultimate aim of reprogramming the microbiome. These aims build on the key contributions we have made to the total synthesis of E. coli genomes as well as applications for the resultant recoded bacteria. In particular, the potential of sense codon reassignment to generate orthogonal genetic communication systems (e.g. viruses cannot infect our cells) provides a powerful approach to address unmet needs for the stability and biosecurity of current gut bacterial models. To this end, as a synthetic genomics group within PNAC we redesign, synthesise, and deploy bacterial species abundant in the microbiome.
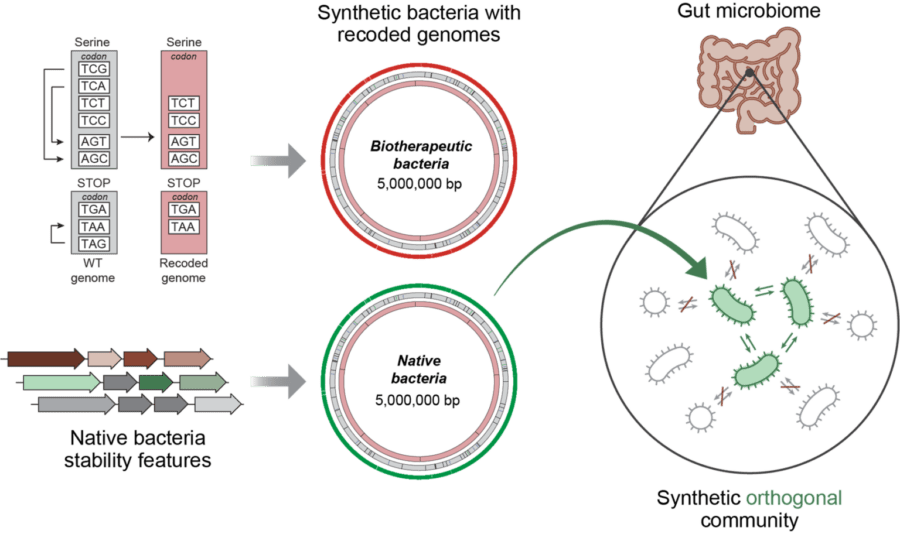
Replacing target codons with synonyms (i.e. recoding), combined with integrating native bacteria stability features, yields synthetic bacterial genome designs with reprogrammed genetic codes that can be deployed in a gut model. These recoded gut bacteria provide a synthetic orthogonal community that is genetically isolated – it does not genetically interact with the endogenous microbiome.
The total synthesis of native bacteria with orthogonal genetic codes, notably, would improve their genetic stability and enable their programmable deployment as biotherapeutics. As a long-term goal, de novo genome design and synthesis technologies will lead to entire synthetic cellular communities that can be used to study and interrogate the microbiome in a biosecure manner.