
All human culture, from society to science, engineering and art, is the product of our brains. And yet, the brain — the biological hardware of cognition — remains enigmatic. To generate new hypotheses about how it works, we need brain maps with an ever-increasing abundance of information.
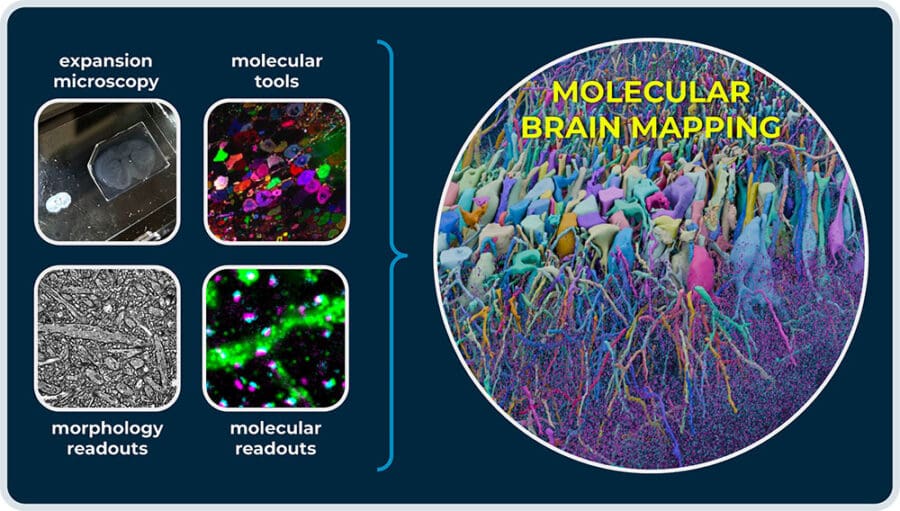
We are generating this abundance of information in our datasets by applying the new paradigm of molecular connectomics. Molecular connectomics combines synaptic resolution circuit mapping with access to the extensive toolboxes of molecular biology to enable multimodal readouts of molecular information:
- In expansion microscopy, we use hydrogels to expand biological tissue by more than 1,000-fold in volume. This separates biological features — increasing resolution — and allows us to map brains at synaptic resolution on conventional light microscopes.
- Light microscopy unlocks the toolboxes of molecular biology, from immunostainings of synaptic markers to RNA cell-typing, viral barcoding of neurons, etc.
- We combine these tools in multimodal multiplexing to read out dozens of targets and different biomolecules within the same brain map.
Our goal is to understand animal behaviour at the molecular level of the biological hardware that gives rise to the computational capabilities of our remarkable brains.
Data credits: E11 Bio: Sung-Yun (Rosa) Park, Arlo Sheridan, William Patton, Julia Lyudchik, Erin Jarvis, Jun Axup, Stephanie Chan, Hugo Damstra, Clarence Magno, Aashir Meeran, Jules Michalska, Michelle Wu, Kathleen Leeper, Sven Truckenbrodt, Johan Winnubst, Andrew Payne. Crick Institute: Samuel Rodriques, Sung-Yun (Rosa) Park. MIT: Ed Boyden, Bobae An, Daniel Leible. MPI: Jörgen Kornfeld, Franz Rieger. 3D rendering: Tyler Sloan.