
Viruses and bacteria are significant causes of human disease. Our group investigates how these pathogens infect their hosts and how hosts attempt to detect and destroy them. The evolutionary arms race between the two continues to provide some of the most unexpected and fascinating discoveries in biology. These insights also drive the development of future medicines.
We take a hypothesis-driven approach and investigate infection from the perspective of both the host and the pathogen. We use a wide range of techniques from biophysics to cellular and in vivo models of infection. In this way, we hope to understand how function is transmitted from molecules to cells.
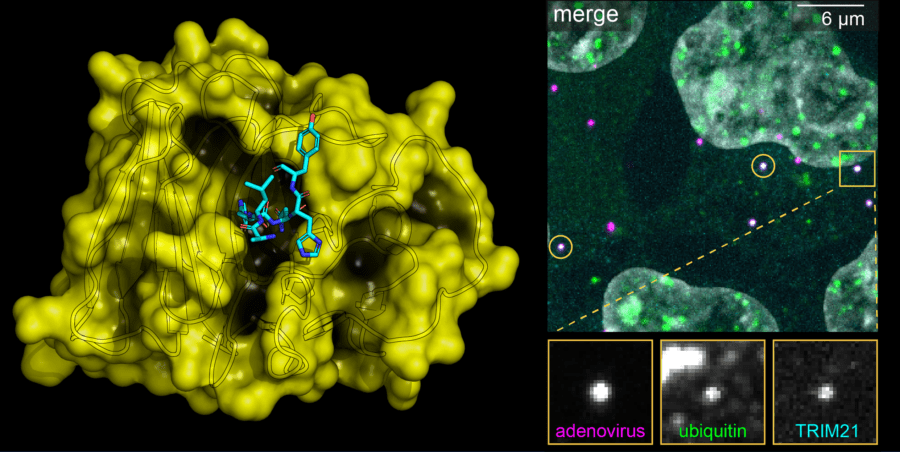
One of our key discoveries was the cytosolic antibody receptor and E3 ligase TRIM21. TRIM21 is expressed in every cell and combines innate and adaptive immune mechanisms to prevent infection. TRIM21 rapidly degrades viruses, proteins and even pathogenic aggregates. We are investigating how TRIM21 works and applying this learning to the development of new technologies and therapeutics. These include the technology Trim-Away, which uses off-the-shelf antibodies to rapidly and specifically deplete proteins, and TRIMTACs – small molecule degraders being developed by spin-out company TRIMTECH Therapeutics.
Another major area of research is the retrovirus HIV-1. We are investigating how HIV-1 builds its capsid and uses it to exploit host machinery while evading host immunity. Recent work includes the discovery that HIV uses dynamic capsid pores to import nucleotides for DNA synthesis and how the cellular metabolite IP6 assembles and stabilises the capsid.