
I started my PhD at the LMB in 1964 under the tutelage of the late John Smith. After this, I spent a couple of years as a postdoc in Jim Watson’s lab at Harvard University, where I co-discovered the first of the RNA polymerase sigma factors.
I returned to the LMB in 1970 as a staff scientist – a position I held until retirement in 2008. At the LMB my work focussed on the organisation and three-dimensional structure of bacterial and eukaryotic chromatin.
We combine topological dynamics and torsional mechanics – essentially spring theory – to study the mechanisms of transcription initiation and elongation by multisubunit RNA polymerases. We model the bacterial enzyme and eukaryotic RNA polymerase II as conical springs. This approach permits not only the assignment of the nature (torsion/tension) and direction of the forces operating during transcriptional processes, but also the timing of their application. When the spring is ‘wound up,’ the molecular assembly possesses potential energy apparent as an altered topology and an associated change in the polypeptide secondary structure. The former can arise from both a suppression of the intrinsic motions of the chain and also from an external force applied by a transcription factor or DNA supercoiling. ‘Unwinding’ does work, and is accompanied by a reversion towards the minimum energy configuration of the chain.
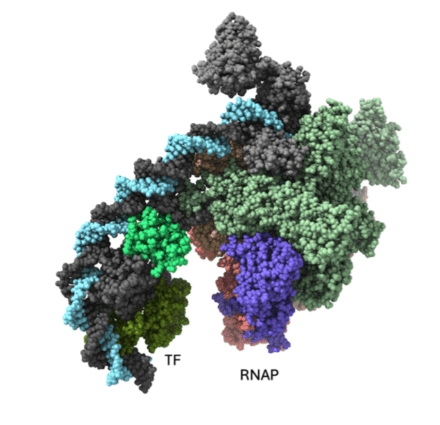
with a transcriptional activator (TF).
Promoter initiation preference is regulated by varying the forces applied to a polymerase assembly, such that the melting of G/C-rich sequences, typical of ribosomal RNA promoters, with high stacking energy is facilitated by torsion-dependent tension, whereas for A/T-rich promoters, less tension is required. Transcription elongation – a low force process – proceeds via an alternation of tensional states pulling the entering DNA into the enzyme one base pair at a time.
This approach can be generalised to other protein assemblies. For example, a chromatin 30 nm fibre can be modelled as a compression spring, while the binding of ATP to the chaperonin GroEL changes the stiffness of the polypeptide chain in the same manner as the RNA polymerase regulators ppGpp and (p)ppApp.