Our group investigates how individual human cells defend themselves against pathogens. We are inspired by the remarkable ability of unicellular organisms to resist infections entirely through cell-autonomous defences. Our goal is to identify principles of cell-autonomous immunity in mammals and how these synergise with professional immune cells. Through an understanding of cell-autonomous immunity, we intend to develop novel strategies against infectious diseases.
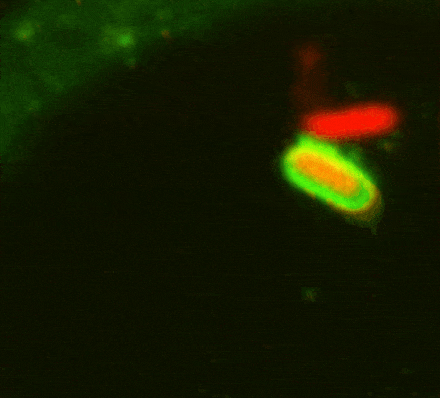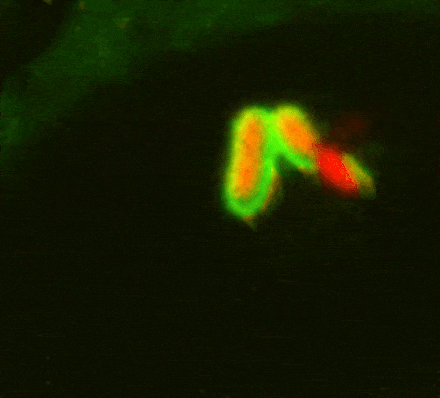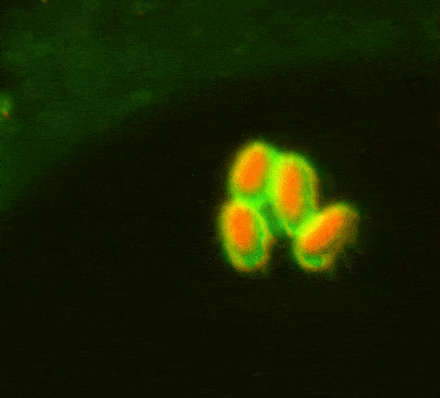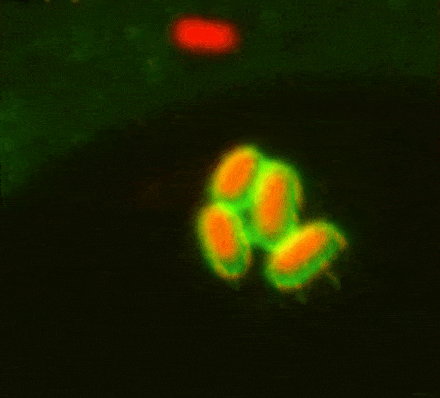
While investigating how human cells protect themselves against cytosol-invading bacteria, we identified two layers of cellular defence. The first involves anti-bacterial autophagy, activated when bacteria damage phagosomal membranes upon entering the host cytosol. This damage exposes sphingomyelin and glycans — molecules normally hidden inside intact phagosomes — for detection by TECPR1 and galectin-8, respectively. We are currently investigating both how sphingomyelin is gradually exposed on stressed but otherwise intact membranes and how its detection by the TECPR1/ATG5/ATG12 complex results in conjugation of mammalian ATG8 to target membranes.
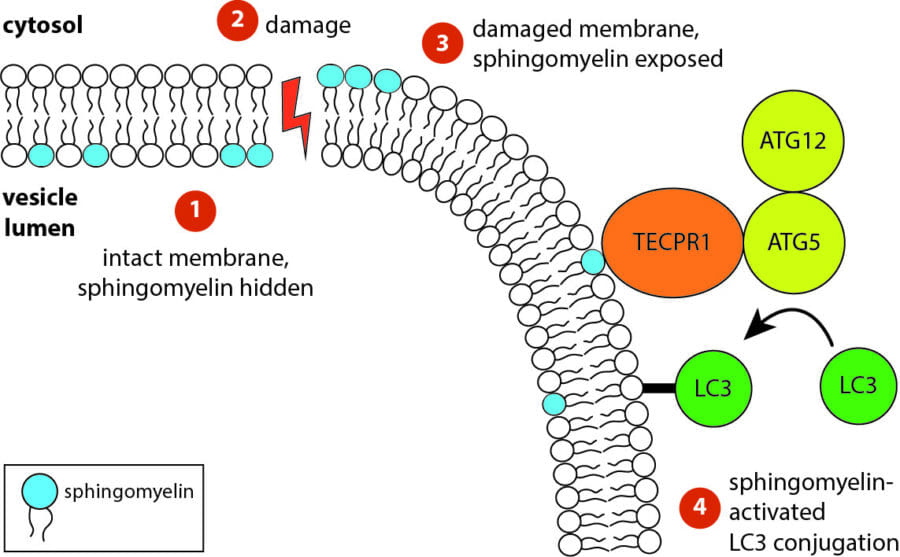
The second line of defence targets bacteria no longer in contact with damaged membranes. These bacteria are transformed into pro-inflammatory and anti-bacterial signalling platforms through cytosolic sensor proteins that trigger the deposition of polyvalent protein coats onto the bacterial surface. Of particular interest to us are the formation of non-canonical inflammasomes triggered by GBP1 and the ubiquitylation of bacterial lipopolysaccharide by RNF213, the first E3 ubiquitin ligase known to target a non-proteinaceous substrate. Ongoing efforts are aimed at understanding how RNF213 restricts pathogens that lack lipopolysaccharide and the identification of RNF213 substrates in non-infected cells.